Page 58 - Read Online
P. 58
Saliba et al. Cancer Drug Resist 2021;4:125-42 I http://dx.doi.org/10.20517/cdr.2020.95 Page 133
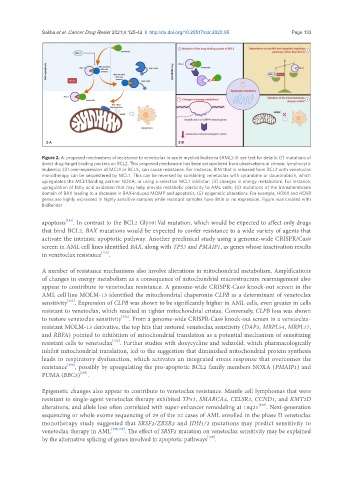
Figure 2. A: proposed mechanisms of resistance to venetoclax in acute myeloid leukemia (AML); B: see text for details: (1) mutations of
direct drug-target binding pockets on BCL2. This proposed mechanism has been extrapolated from observations in chronic lymphocytic
leukemia; (2) over-expression of MCL1 or BCLX L can cause resistance. For instance, BIM that is released from BCL2 with venetoclax
monotherapy can be sequestered by MCL1. This can be reversed by combining venetoclax with cytarabine or daunorubicin, which
upregulates the MCL1 binding partner NOXA, or using a selective MCL1 inhibitor; (3) changes in energy metabolism. For instance,
upregulation of fatty acid oxidation that may help provide metabolic plasticity to AML cells; (4) mutations of the transmembrane
domain of BAX leading to a decrease in BAX-induced MOMP and apoptosis; (5) epigenetic alterations. For example, HOXA and HOXB
genes are highly expressed in highly sensitive samples while resistant samples have little or no expression. Figure was created with
BioRender
apoptosis [154] . In contrast to the BCL2 Gly101Val mutation, which would be expected to affect only drugs
that bind BCL2, BAX mutations would be expected to confer resistance to a wide variety of agents that
activate the intrinsic apoptotic pathway. Another preclinical study using a genome-wide CRISPR/Cas9
screen in AML cell lines identified BAX, along with TP53 and PMAIP1, as genes whose inactivation results
in venetoclax resistance [155] .
A number of resistance mechanisms also involve alterations in mitochondrial metabolism. Amplifications
of changes in energy metabolism as a consequence of mitochondrial macrostructure rearrangement also
appear to contribute to venetoclax resistance. A genome-wide CRISPR-Cas9 knock-out screen in the
AML cell line MOLM-13 identified the mitochondrial chaperonin CLPB as a determinant of venetoclax
sensitivity [132] . Expression of CLPB was shown to be significantly higher in AML cells, even greater in cells
resistant to venetoclax, which resulted in tighter mitochondrial cristae. Conversely, CLPB loss was shown
to restore venetoclax sensitivity [132] . From a genome-wide CRISPR-Cas9 knock-out screen in a venetoclax-
resistant MOLM-13 derivative, the top hits that restored venetoclax sensitivity (DAP3, MRPL54, MRPL17,
and RBFA) pointed to inhibition of mitochondrial translation as a potential mechanism of sensitizing
resistant cells to venetoclax [156] . Further studies with doxycycline and tedizolid, which pharmacologically
inhibit mitochondrial translation, led to the suggestion that diminished mitochondrial protein synthesis
leads to respiratory dysfunction, which activates an integrated stress response that overcomes the
resistance [156] , possibly by upregulating the pro-apoptotic BCL2 family members NOXA (PMAIP1) and
PUMA (BBC3) [157] .
Epigenetic changes also appear to contribute to venetoclax resistance. Mantle cell lymphomas that were
resistant to single-agent venetoclax therapy exhibited TP53, SMARCA4, CELSR3, CCND1, and KMT2D
alterations, and allele loss often correlated with super-enhancer remodeling at 18q21 [158] . Next-generation
sequencing or whole exome sequencing of 29 of the 32 cases of AML enrolled in the phase II venetoclax
monotherapy study suggested that SRSF2/ZRSR2 and IDH1/2 mutations may predict sensitivity to
venetoclax therapy in AML [138,159] . The effect of SRSF2 mutation on venetoclax sensitivity may be explained
by the alternative splicing of genes involved in apoptotic pathways [160] .