Page 57 - Read Online
P. 57
Elton et al. Cancer Drug Resist 2020;3:161-70 I http://dx.doi.org/10.20517/cdr.2019.117 Page 167
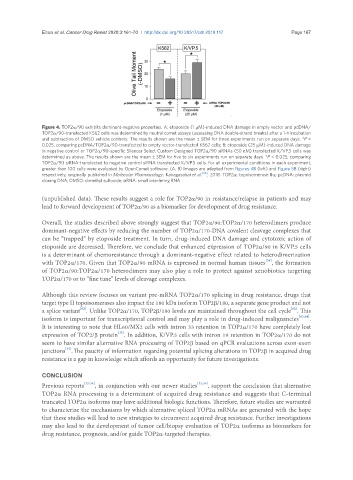
Figure 4. TOP2α/90 exhibits dominant-negative properties. A: etoposide (1 µM)-induced DNA damage in empty vector and pcDNA/
TOP2α/90-transfected K562 cells was determined by neutral comet assays (assessing DNA double-strand breaks) after a 1-h incubation
and subtraction of DMSO vehicle controls. The results shown are the mean ± SEM for three experiments run on separate days. *P <
0.025, comparing pcDNA/TOP2α/90-transfected to empty vector-transfected K562 cells; B: etoposide (25 µM)-induced DNA damage
in negative control or TOP2α/90-specific Silencer Select Custom Designed TOP2α/90 siRNAs (50 nM) transfected K/VP.5 cells was
determined as above. The results shown are the mean ± SEM for five to six experiments run on separate days. *P < 0.025, comparing
TOP2α/90 siRNA-transfected to negative control siRNA transfected K/VP.5 cells. For all experimental conditions in each experiment,
greater than 100 cells were evaluated by OpenComet software. (A, B) Images are adapted from Figures 4B (left) and Figure 5B (right)
respectively, originally published in Molecular Pharmacology; Kanagasabai et al. [36] , 2018. TOP2α: topoisomerase IIα; pcDNA: plasmid
cloning DNA; DMSO: dimethyl sulfoxide; siRNA: small interfering RNA
(unpublished data). These results suggest a role for TOP2α/90 in resistance/relapse in patients and may
lead to forward development of TOP2α/90 as a biomarker for development of drug resistance.
Overall, the studies described above strongly suggest that TOP2α/90:TOP2α/170 heterodimers produce
dominant-negative effects by reducing the number of TOP2α/170-DNA covalent cleavage complexes that
can be “trapped” by etoposide treatment. In turn, drug-induced DNA damage and cytotoxic action of
etoposide are decreased. Therefore, we conclude that enhanced expression of TOP2α/90 in K/VP.5 cells
is a determinant of chemoresistance through a dominant-negative effect related to heterodimerization
[36]
with TOP2α/170. Given that TOP2α/90 mRNA is expressed in normal human tissues , the formation
of TOP2α/90:TOP2α/170 heterodimers may also play a role to protect against xenobiotics targeting
TOP2α/170 or to “fine tune” levels of cleavage complexes.
Although this review focuses on variant pre-mRNA TOP2α/170 splicing in drug resistance, drugs that
target type II topoisomerases also impact the 180 kDa isoform TOP2β/180, a separate gene product and not
[62]
[62]
a splice variant . Unlike TOP2α/170, TOP2β/180 levels are maintained throughout the cell cycle . This
isoform is important for transcriptional control and may play a role in drug-induced malignancies [63,64] .
It is interesting to note that HL60/MX2 cells with intron 33 retention in TOP2α/170 have completely lost
[32]
expression of TOP2/β protein . In addition, K/VP.5 cells with intron 19 retention in TOP2α/170 do not
seem to have similar alternative RNA processing of TOP2β based on qPCR evaluations across exon-exon
[35]
junctions .The paucity of information regarding potential splicing alterations in TOP2β in acquired drug
resistance is a gap in knowledge which affords an opportunity for future investigations.
CONCLUSION
Previous reports [32-34] , in conjunction with our newer studies [35,36] , support the conclusion that alternative
TOP2α RNA processing is a determinant of acquired drug resistance and suggests that C-terminal
truncated TOP2α isoforms may have additional biologic functions. Therefore, future studies are warranted
to characterize the mechanisms by which alternative spliced TOP2α mRNAs are generated with the hope
that these studies will lead to new strategies to circumvent acquired drug resistance. Further investigations
may also lead to the development of tumor cell/biopsy evaluation of TOP2α isoforms as biomarkers for
drug resistance, prognosis, and/or guide TOP2α-targeted therapies.