Page 43 - Read Online
P. 43
Lotz et al. Cancer Drug Resist 2020;3:149-60 I http://dx.doi.org/10.20517/cdr.2019.114 Page 153
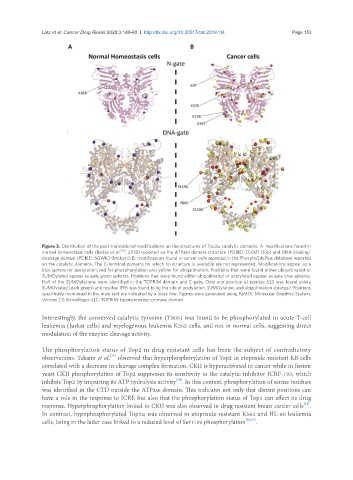
Figure 3. Distribution of the post-translational modifications on the structures of Top2a catalytic domains. A: modifications found in
normal homeostasis cells (Bedez et al. [16] , 2018) reported on the ATPase domain structure (PDBID: 1ZXM) (top) and DNA binding/
cleavage domain (PDBID: 5GWK) (bottom). B: modifications found in cancer cells reported in the PhosphoSitePlus database reported
on the catalytic domains. The C-terminal domains for which no structure is available are not represented. Modifications appear as a
blue sphere for acetylation, red for phosphorylation and yellow for ubiquitination. Positions that were found either ubiquitinated or
SUMOylated appear as pale green spheres. Positions that were found either ubiquitinated or acetylated appear as pale blue spheres.
Half of the SUMOylations were identified in the TOPRIM domain and C-gate. Only one position at residue 625 was found solely
SUMOylated (dark green) and residue 1196 was found to be the site of acetylation, SUMOylation, and ubiquitination (orange). Positions
specifically mentioned in the main text are indicated by a black line. Figures were generated using PyMOL Molecular Graphics System,
Version 2.0 Schrödinger, LLC. TOPRIM: topoisomerase-primase domain
Interestingly, the conserved catalytic tyrosine (Y805) was found to be phosphorylated in acute T-cell
leukemia (Jurkat cells) and myelogenous leukemia K562 cells, and not in normal cells, suggesting direct
modulation of the enzyme cleavage activity.
The phosphorylation status of Top2 in drug resistant cells has been the subject of contradictory
[33]
observations. Takano et al. observed that hyperphosphorylation of Top2 in etoposide-resistant KB cells
correlated with a decrease in cleavage complex formation. CKII is hyperactivated in cancer while in fission
yeast CKII phosphorylation of Top2 suppresses its sensitivity to the catalytic inhibitor ICRF-193, which
[34]
inhibits Top2 by impairing its ATP hydrolysis activity . In this context, phosphorylation of serine residues
was identified in the CTD outside the ATPase domain. This indicates not only that distant positions can
have a role in the response to ICRF, but also that the phosphorylation status of Top2 can affect its drug
[35]
response. Hyperphosphorylation linked to CKII was also observed in drug resistant breast cancer cells .
In contrast, hypophosphorylated Top2a was observed in etoposide-resistant K562 and HL-60 leukemia
cells, being in the latter case linked to a reduced level of Ser1106 phosphorylation [36,37] .