Page 14 - Read Online
P. 14
Page 996 Gmeiner. Cancer Drug Resist 2019;2:994-1001 I http://dx.doi.org/10.20517/cdr.2019.95
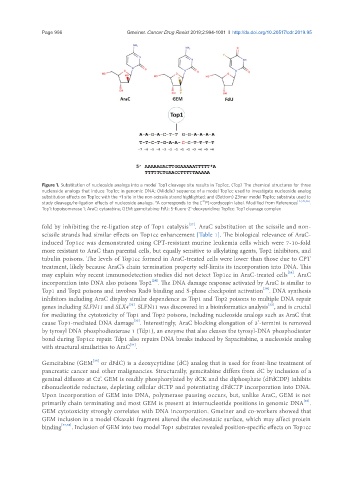
Figure 1. Substitution of nucleoside analogs into a model Top1 cleavage site results in Top1cc. (Top) The chemical structures for three
nucleoside analogs that induce Top1cc in genomic DNA; (Middle) sequence of a model Top1cc used to investigate nucleoside analog
substitution effects on Top1cc with the +1 site in the non-scissile strand highlighted; and (Bottom) 23mer model Top1cc substrate used to
32
study cleavage/re-ligation effects of nucleoside analogs. *A corresponds to the [ P]-cordycepin label. Modified from References [27,39,46] .
Top1: topoisomerase 1; AraC: cytarabine; GEM: gemcitabine; FdU: 5-fluoro-2’-deoxyuridine; Top1cc: Top1 cleavage complex
[27]
fold by inhibiting the re-ligation step of Top1 catalysis . AraC substitution at the scissile and non-
scissile strands had similar effects on Top1cc enhancement [Table 1]. The biological relevance of AraC-
induced Top1cc was demonstrated using CPT-resistant murine leukemia cells which were 7-10-fold
more resistant to AraC than parental cells, but equally sensitive to alkylating agents, Top2 inhibitors, and
tubulin poisons. The levels of Top1cc formed in AraC-treated cells were lower than those due to CPT
treatment, likely because AraC’s chain termination property self-limits its incorporation into DNA. This
[28]
may explain why recent immunodetection studies did not detect Top1cc in AraC-treated cells . AraC
[29]
incorporation into DNA also poisons Top2 . The DNA damage response activated by AraC is similar to
[30]
Top1 and Top2 poisons and involves Rad9 binding and S-phase checkpoint activation . DNA synthesis
inhibitors including AraC display similar dependence as Top1 and Top2 poisons to multiple DNA repair
[31]
[32]
genes including SLFN11 and SLX4 . SLFN11 was discovered in a bioinformatics analysis , and is crucial
for mediating the cytotoxicity of Top1 and Top2 poisons, including nucleoside analogs such as AraC that
[33]
cause Top1-mediated DNA damage . Interestingly, AraC blocking elongation of 3’-termini is removed
by tyrosyl DNA phosphodiesterase 1 (Tdp1), an enzyme that also cleaves the tyrosyl-DNA phosphodiester
bond during Top1cc repair. Tdp1 also repairs DNA breaks induced by Sapacitabine, a nucleoside analog
[34]
with structural similarities to AraC .
[35]
Gemcitabine (GEM or dFdC) is a deoxycytidine (dC) analog that is used for front-line treatment of
pancreatic cancer and other malignancies. Structurally, gemcitabine differs from dC by inclusion of a
geminal difluoro at C2’. GEM is readily phosphorylated by dCK and the diphosphate (dFdCDP) inhibits
ribonucleotide reductase, depleting cellular dCTP and potentiating dFdCTP incorporation into DNA.
Upon incorporation of GEM into DNA, polymerase pausing occurs, but, unlike AraC, GEM is not
[36]
primarily chain terminating and most GEM is present at internucleotide positions in genomic DNA .
GEM cytotoxicity strongly correlates with DNA incorporation. Gmeiner and co-workers showed that
GEM inclusion in a model Okazaki fragment altered the electrostatic surface, which may affect protein
binding [37,38] . Inclusion of GEM into two model Top1 substrates revealed position-specific effects on Top1cc