Page 61 - Read Online
P. 61
Meyer et al. Cancer Drug Resist 2019;2:313-25 I http://dx.doi.org/10.20517/cdr.2019.11 Page 315
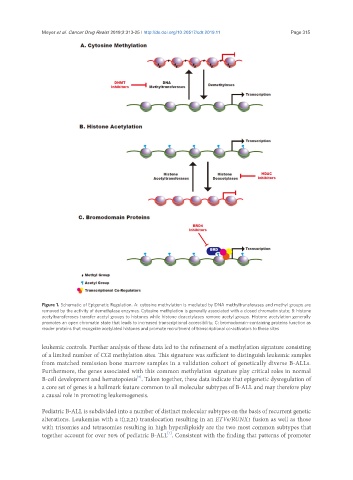
Figure 1. Schematic of Epigenetic Regulation. A: cytosine methylation is mediated by DNA methyltransferases and methyl groups are
removed by the activity of demethylase enzymes. Cytosine methylation is generally associated with a closed chromatin state; B: histone
acetyltransferases transfer acetyl groups to histones while histone deacetylases remove acetyl groups. Histone acetylation generally
promotes an open chromatin state that leads to increased transcriptional accessibility; C: bromodomain-containing proteins function as
reader proteins that recognize acetylated histones and promote recruitment of transcriptional co-activators to those sites
leukemic controls. Further analysis of these data led to the refinement of a methylation signature consisting
of a limited number of CGI methylation sites. This signature was sufficient to distinguish leukemic samples
from matched remission bone marrow samples in a validation cohort of genetically diverse B-ALLs.
Furthermore, the genes associated with this common methylation signature play critical roles in normal
[9]
B-cell development and hematopoiesis . Taken together, these data indicate that epigenetic dysregulation of
a core set of genes is a hallmark feature common to all molecular subtypes of B-ALL and may therefore play
a causal role in promoting leukemogenesis.
Pediatric B-ALL is subdivided into a number of distinct molecular subtypes on the basis of recurrent genetic
alterations. Leukemias with a t(12;21) translocation resulting in an ETV6/RUNX1 fusion as well as those
with trisomies and tetrasomies resulting in high hyperdiploidy are the two most common subtypes that
[1]
together account for over 50% of pediatric B-ALL . Consistent with the finding that patterns of promoter