Page 139 - Read Online
P. 139
Murthy et al. Cancer Drug Resist 2019;2:665-79 I http://dx.doi.org/10.20517/cdr.2019.002 Page 673
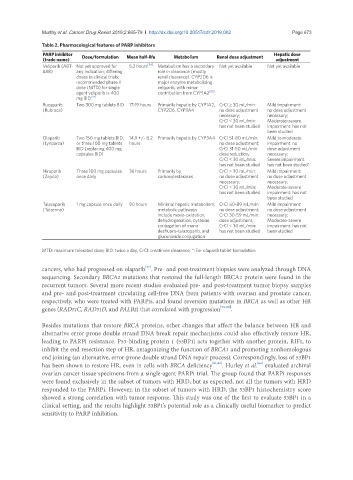
Table 2. Pharmacological features of PARP inhibitors
PARP inhibitor Dose/formulation Mean half-life Metabolism Renal dose adjustment Hepatic dose
(trade name) adjustment
Veliparib (ABT- Not yet approved for 5.2 hours [54] Metabolism has a secondary Not yet available Not yet available
888) any indication; differing role in clearance (mostly
doses in clinical trials; renal clearance). CYP2D6 is
recommended phase II major enzyme metabolizing
dose (MTD) for single veliparib, with minor
agent veliparib is 400 contribution from CYP1A2 [55]
mg BID [54]
Rucaparib Two 300 mg tablets BID 17-19 hours Primarily hepatic by CYP1A2, CrCl ≥ 30 mL/min: Mild impairment:
(Rubraca) CYP2D6, CYP3A4 no dose adjustment no dose adjustment
necessary; necessary;
CrCl < 30 mL/min: Moderate-severe
has not been studied impairment: has not
been studied
Olaparib Two 150 mg tablets BID, 14.9 +/- 8.2 Primarily hepatic by CYP3A4 CrCl 51-80 mL/min: Mild to moderate
(Lynparza) or three 100 mg tablets hours no dose adjustment; impairment: no
BID (replacing 400 mg CrCl 31-50 mL/min: dose adjustment
capsules BID) dose reduction; necessary;
CrCl < 30 mL/min: Severe impairment:
has not been studied has not been studied*
Niraparib Three 100 mg capsules 36 hours Primarily by CrCl ≥ 30 mL/min: Mild impairment:
(Zejula) once daily carboxylesterases no dose adjustment no dose adjustment
necessary; necessary;
CrCl < 30 mL/min: Moderate-severe
has not been studied impairment: has not
been studied
Talazoparib 1 mg capsule once daily 90 hours Minimal hepatic metabolism; CrCl 60-89 mL/min: Mild impairment:
(Talzenna) metabolic pathways no dose adjustment; no dose adjustment
include mono-oxidation, CrCl 30-59 mL/min: necessary;
dehydrogenation, cysteine dose adjustment; Moderate-severe
conjugation of mono- CrCl < 30 mL/min: impairment: has not
desfluoro-talazoparib, and has not been studied been studied
glucuronide conjugation
MTD: maximum tolerated dose; BID: twice a day; CrCl: creatinine clearance; *: For olaparib tablet formulation
[57]
cancers, who had progressed on olaparib . Pre- and post-treatment biopsies were analyzed through DNA
sequencing. Secondary BRCA2 mutations that restored the full-length BRCA2 protein were found in the
recurrent tumors. Several more recent studies evaluated pre- and post-treatment tumor biopsy samples
and pre- and post-treatment circulating cell-free DNA from patients with ovarian and prostate cancer,
respectively, who were treated with PARPis, and found reversion mutations in BRCA as well as other HR
genes (RAD51C, RAD51D, and PALB2) that correlated with progression [58-60] .
Besides mutations that restore BRCA proteins, other changes that affect the balance between HR and
alternative error-prone double strand DNA break repair mechanisms could also effectively restore HR,
leading to PARPi resistance. P53-binding protein 1 (53BP1) acts together with another protein, RIF1, to
inhibit the end resection step of HR, antagonizing the function of BRCA1 and promoting nonhomologous
end joining (an alternative, error-prone double strand DNA repair process). Correspondingly, loss of 53BP1
[63]
has been shown to restore HR, even in cells with BRCA deficiency [61,62] . Hurley et al. evaluated archival
ovarian cancer tissue specimens from a single-agent PARPi trial. The group found that PARPi responses
were found exclusively in the subset of tumors with HRD, but as expected, not all the tumors with HRD
responded to the PARPi. However, in the subset of tumors with HRD, the 53BP1 histochemistry score
showed a strong correlation with tumor response. This study was one of the first to evaluate 53BP1 in a
clinical setting, and the results highlight 53BP1’s potential role as a clinically useful biomarker to predict
sensitivity to PARP inhibition.