Page 140 - Read Online
P. 140
Maiocchi et al. Vessel Plus 2023;7:27 https://dx.doi.org/10.20517/2574-1209.2023.69 Page 3 of 19
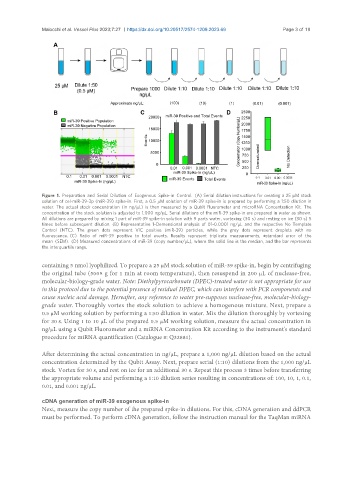
Figure 1. Preparation and Serial Dilution of Exogenous Spike-in Control. (A) Serial dilution instructions for creating a 25 μM stock
solution of cel-miR-39-3p (miR-39) spike-in. First, a 0.5 μM solution of miR-39 spike-in is prepared by performing a 1:50 dilution in
water. The actual stock concentration (in ng/μL) is then measured by a Qubit Fluorometer and microRNA Concentration Kit. The
concentration of the stock solution is adjusted to 1,000 ng/μL. Serial dilutions of the miR-39 spike-in are prepared in water as shown.
All dilutions are prepared by mixing 1 part of miR-39 spike-in solution with 9 parts water, vortexing (30 s) and resting on ice (30 s) 5
times before subsequent dilution. (B) Representative 1-Demensional analysis of 01-0.0001 ng/μL and the respective No Template
Control (NTC). The green dots represent VIC positive (miR-39) particles, while the grey dots represent droplets with no
fluorescence. (C) Ratio of miR-39 positive to total events. Results represent triplicate measurements, ±standard error of the
mean (SEM). (D) Measured concentrations of miR-39 (copy number/μL), where the solid line is the median, and the bar represents
the interquartile range.
containing 5 nmol lyophilized. To prepare a 25 µM stock solution of miR-39 spike-in, begin by centrifuging
the original tube (500× g for 1 min at room temperature), then resuspend in 200 µL of nuclease-free,
molecular-biology-grade water. Note: Diethylpyrocarbonate (DPEC)-treated water is not appropriate for use
in this protocol due to the potential presence of residual DPEC, which can interfere with PCR components and
cause nucleic acid damage. Hereafter, any reference to water pre-supposes nuclease-free, molecular-biology-
grade water. Thoroughly vortex the stock solution to achieve a homogenous mixture. Next, prepare a
0.5 µM working solution by performing a 1:50 dilution in water. Mix the dilution thoroughly by vortexing
for 30 s. Using 1 to 10 µL of the prepared 0.5 µM working solution, measure the actual concentration in
ng/µL using a Qubit Fluorometer and a miRNA Concentration Kit according to the instrument's standard
procedure for miRNA quantification (Catalogue #: Q32881).
After determining the actual concentration in ng/µL, prepare a 1,000 ng/µL dilution based on the actual
concentration determined by the Qubit Assay. Next, prepare serial (1:10) dilutions from the 1,000 ng/µL
stock. Vortex for 30 s, and rest on ice for an additional 30 s. Repeat this process 3 times before transferring
the appropriate volume and performing a 1:10 dilution series resulting in concentrations of: 100, 10, 1, 0.1,
0.01, and 0.001 ng/µL.
cDNA generation of miR-39 exogenous spike-in
Next, measure the copy number of the prepared spike-in dilutions. For this, cDNA generation and ddPCR
must be performed. To perform cDNA generation, follow the instruction manual for the TaqMan miRNA