Page 113 - Read Online
P. 113
Almurshidi et al. Neuroimmunol Neuroinflammation 2019;6:11 I http://dx.doi.org/10.20517/2347-8659.2019.19 Page 3 of 11
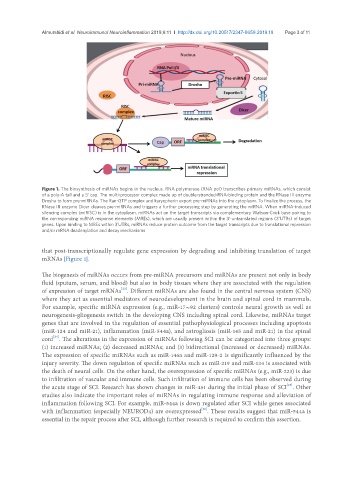
Figure 1. The biosynthesis of miRNAs begins in the nucleus. RNA polymerase (RNA pol) transcribes primary miRNAs, which consist
of a poly-A tail and a 5’ cap. The multi-processor complex made up of double-stranded-RNA-binding protein and the RNase III enzyme
Drosha to form pre-miRNAs. The Ran-GTP complex and karyopherin export pre-miRNAs into the cytoplasm. To finalize the process, the
RNase III enzyme Dicer cleaves pre-miRNAs and triggers a further processing step by generating the miRNA. When miRNA-induced
silencing complex (miRISC) is in the cytoplasm, miRNAs act on the target transcripts via complementary Watson-Crick base pairing to
the corresponding miRNA response elements (MREs), which are usually present within the 3′-untranslated regions (3’UTRs) of target
genes. Upon binding to MREs within 3’UTRs, miRNAs reduce protein outcome from the target transcripts due to translational repression
and/or mRNA deadenylation and decay mechanisms
that post-transcriptionally regulate gene expression by degrading and inhibiting translation of target
mRNAs [Figure 1].
The biogenesis of miRNAs occurs from pre-miRNA precursors and miRNAs are present not only in body
fluid (sputum, serum, and blood) but also in body tissues where they are associated with the regulation
[23]
of expression of target mRNAs . Different miRNAs are also found in the central nervous system (CNS)
where they act as essential mediators of neurodevelopment in the brain and spinal cord in mammals.
For example, specific miRNA expression (e.g., miR-17∼92 clusters) controls neural growth as well as
neurogenesis-gliogenesis switch in the developing CNS including spinal cord. Likewise, miRNAs target
genes that are involved in the regulation of essential pathophysiological processes including apoptosis
(miR-124 and miR-21), inflammation (miR-544a), and astrogliosis (miR-145 and miR-21) in the spinal
[24]
cord . The alterations in the expression of miRNAs following SCI can be categorized into three groups:
(1) increased miRNAs; (2) decreased miRNAs; and (3) bidirectional (increased or decreased) miRNAs.
The expression of specific miRNAs such as miR-146a and miR-129-2 is significantly influenced by the
injury severity. The down regulation of specific miRNAs such as miR-219 and miR-124 is associated with
the death of neural cells. On the other hand, the overexpression of specific miRNAs (e.g., miR-223) is due
to infiltration of vascular and immune cells. Such infiltration of immune cells has been observed during
[25]
the acute stage of SCI. Research has shown changes in miR-451 during the initial phase of SCI . Other
studies also indicate the important roles of miRNAs in regulating immune response and alleviation of
inflammation following SCI. For example, miR-544a is down regulated after SCI while genes associated
with inflammation (especially NEUROD4) are overexpressed . These results suggest that miR-544a is
[26]
essential in the repair process after SCI, although further research is required to confirm this assertion.