Page 28 - Read Online
P. 28
Agresti et al. J Transl Genet Genom 2018;2:9 I http://dx.doi.org/10.20517/jtgg.2018.05 Page 3 of 11
Table 1. Notable mitochondrial functions with its associated purpose (in addition to oxidative phosphorylation)
Mitochondrial function Purpose
Formation of Iron-Sulfur (Fe-S) clusters [2] Small prosthetic groups that are inorganic and composed of iron-sulfur clusters [9]
Role in electron transfer, binding and activation of substrates, redox catalysis, DNA replication,
DNA repair, gene expression regulation, and modification of tRNAs [9]
β-oxidation of fatty acids [2] Functions to drive oxidative phosphorylation and supply acetyl-CoA for ketogenesis [10]
Synthesis of heme prosthetic groups [2] Production of heme through a complex biochemical process occurring in cells [11]
Steroidogenesis [2] Production of steroid hormones from cholesterol [12]
Urea cycle [2] Production of urea from ammonia [13]
Homeostatic maintenance of calcium [2] Responsible for maintenance of calcium homeostasis required for biological processes [14]
Generation of free radicals [4] Production of toxic byproducts resultant from mitochondrial energetic processes (i.e., oxidative
phosphorylation) [15]
Inflammation and innate immunity [4] Mitochondria serve as a hub for innate immune signaling [16]
Mitochondrial damage leads to inflammation by releasing mitochondrial alarmins [16]
Cellular signals associated with Mitochondrial participation in the regulation of cell survival and cell death induced by oxidative
[17]
cell survival and death [2] stress
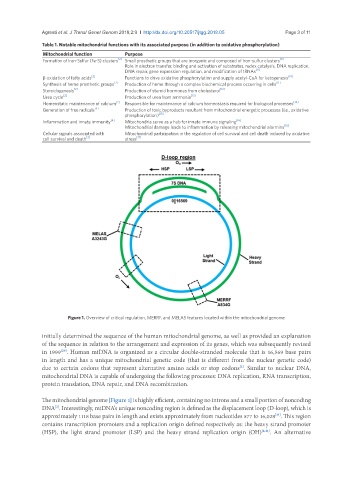
Figure 1. Overview of critical regulation, MERRF, and MELAS features located within the mitochondrial genome
initially determined the sequence of the human mitochondrial genome, as well as provided an explanation
of the sequence in relation to the arrangement and expression of its genes, which was subsequently revised
in 1999 . Human mtDNA is organized as a circular double-stranded molecule that is 16,569 base pairs
[20]
in length and has a unique mitochondrial genetic code (that is different from the nuclear genetic code)
due to certain codons that represent alternative amino acids or stop codons . Similar to nuclear DNA,
[2]
mitochondrial DNA is capable of undergoing the following processes: DNA replication, RNA transcription,
protein translation, DNA repair, and DNA recombination.
The mitochondrial genome [Figure 1] is highly efficient, containing no introns and a small portion of noncoding
DNA . Interestingly, mtDNA’s unique noncoding region is defined as the displacement loop (D-loop), which is
[2]
approximately 1118 base pairs in length and exists approximately from nucleotides 577 to 16,028 . This region
[21]
contains transcription promoters and a replication origin defined respectively as: the heavy strand promoter
(HSP), the light strand promoter (LSP) and the heavy strand replication origin (OH) [2,21] . An alternative