Page 76 - Read Online
P. 76
Page 70 Leung et al. J Transl Genet Genom 2023;7:79-86 https://dx.doi.org/10.20517/jtgg.2023.09
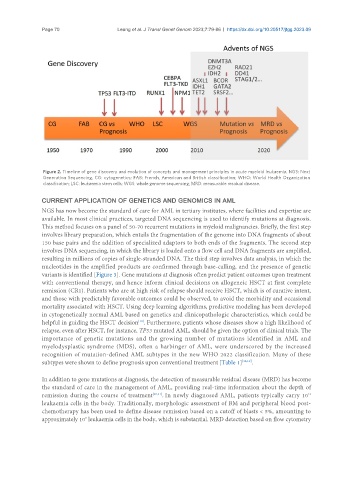
Figure 2. Timeline of gene discovery and evolution of concepts and management principles in acute myeloid leukaemia. NGS: Next
Generation Sequencing, CG: cytogenetics; FAB: French, American and British classification; WHO: World Health Organization
classification; LSC: leukaemia stem cells; WGS: whole genome sequencing; MRD: measurable residual disease.
CURRENT APPLICATION OF GENETICS AND GENOMICS IN AML
NGS has now become the standard of care for AML in tertiary institutes, where facilities and expertise are
available. In most clinical practices, targeted DNA sequencing is used to identify mutations at diagnosis.
This method focuses on a panel of 50-70 recurrent mutations in myeloid malignancies. Briefly, the first step
involves library preparation, which entails the fragmentation of the genome into DNA fragments of about
150 base pairs and the addition of specialized adaptors to both ends of the fragments. The second step
involves DNA sequencing, in which the library is loaded onto a flow cell and DNA fragments are amplified,
resulting in millions of copies of single-stranded DNA. The third step involves data analysis, in which the
nucleotides in the amplified products are confirmed through base-calling, and the presence of genetic
variants is identified [Figure 3]. Gene mutations at diagnosis often predict patient outcomes upon treatment
with conventional therapy, and hence inform clinical decisions on allogeneic HSCT at first complete
remission (CR1). Patients who are at high risk of relapse should receive HSCT, which is of curative intent,
and those with predictably favorable outcomes could be observed, to avoid the morbidity and occasional
mortality associated with HSCT. Using deep learning algorithms, predictive modeling has been developed
in cytogenetically normal AML based on genetics and clinicopathologic characteristics, which could be
helpful in guiding the HSCT decision . Furthermore, patients whose diseases show a high likelihood of
[18]
relapse, even after HSCT, for instance, TP53 mutated AML, should be given the option of clinical trials. The
importance of genetic mutations and the growing number of mutations identified in AML and
myelodysplastic syndrome (MDS), often a harbinger of AML, were underscored by the increased
recognition of mutation-defined AML subtypes in the new WHO 2022 classification. Many of these
subtypes were shown to define prognosis upon conventional treatment [Table 1] [12,19] .
In addition to gene mutations at diagnosis, the detection of measurable residual disease (MRD) has become
the standard of care in the management of AML, providing real-time information about the depth of
remission during the course of treatment [20,21] . In newly diagnosed AML, patients typically carry 10
12
leukaemia cells in the body. Traditionally, morphologic assessment of BM and peripheral blood post-
chemotherapy has been used to define disease remission based on a cutoff of blasts < 5%, amounting to
approximately 10 leukaemia cells in the body, which is substantial. MRD detection based on flow cytometry
9