Page 78 - Read Online
P. 78
Guerriero et al. Hepatoma Res 2019;5:6 I http://dx.doi.org/10.20517/2394-5079.2018.108 Page 11 of 22
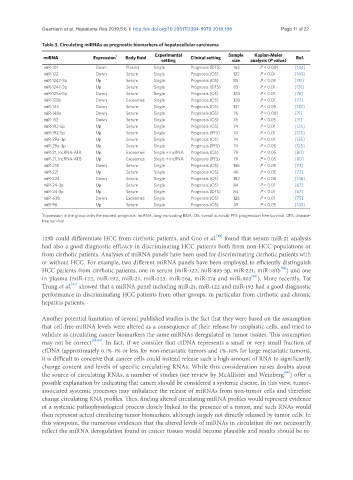
Table 3. Circulating miRNAs as prognostic biomarkers of hepatocellular carcinoma
Experimental Sample Kaplan-Meier
miRNA Expression 1 Body fluid Clinical setting Ref.
setting size analysis (P value)
miR-101 Down Plasma Single Prognosis (DFS) 163 P < 0.001 [144]
miR-122 Down Serum Single Prognosis (OS) 122 P < 0.01 [145]
miR-1247-3p Up Serum Single Prognosis (OS) 85 P < 0.05 [131]
miR-1247-3p Up Serum Single Prognosis (DFS) 85 P < 0.01 [131]
miR-125a-5p Down Serum Single Prognosis (OS) 120 P < 0.01 [76]
miR-125b Down Exosomes Single Prognosis (OS) 128 P < 0.01 [77]
miR-143 Down Serum Single Prognosis (OS) 131 P < 0.05 [132]
miR-148a Down Serum Single Prognosis (OS) 76 P < 0.001 [71]
miR-152 Down Serum Single Prognosis (OS) 76 P < 0.05 [71]
miR-192-5p Up Serum Single Prognosis (OS) 74 P < 0.01 [125]
miR-192-5p Up Serum Single Prognosis (PFS) 74 P < 0.01 [125]
miR-29a-3p Up Serum Single Prognosis (OS) 74 P < 0.01 [125]
miR-29a-3p Up Serum Single Prognosis (PFS) 74 P < 0.05 [125]
miR-21, lncRNA-ATB Up Exosomes Single + lncRNA Prognosis (OS) 79 P < 0.05 [80]
miR-21, lncRNA-ATB Up Exosomes Single + lncRNA Prognosis (PFS) 79 P < 0.05 [80]
miR-218 Down Serum Single Prognosis (OS) 156 P < 0.05 [73]
miR-221 Up Serum Single Prognosis (OS) 46 P < 0.05 [72]
miR-224 Down Serum Single Prognosis (OS) 182 P < 0.05 [146]
miR-24-3p Up Serum Single Prognosis (OS) 84 P < 0.01 [67]
miR-24-3p Up Serum Single Prognosis (DFS) 84 P < 0.01 [67]
miR-638 Down Exosomes Single Prognosis (OS) 126 P < 0.01 [75]
miR-96 Up Serum Single Prognosis (OS) 49 P < 0.05 [143]
1 Expression in the group with the poorest prognosis. lncRNA: long noncoding RNA; OS: overall survival; PFS: progression free survival; DFS: disease-
free survival
[59]
125b could differentiate HCC from cirrhotic patients, and Guo et al. found that serum miR-21 analysis
had also a good diagnostic efficacy in discriminating HCC patients both from non-HCC populations or
from cirrhotic patients. Analyses of miRNA panels have been used for discriminating cirrhotic patients with
or without HCC. For example, two different miRNA panels have been employed to efficiently distinguish
[60]
HCC patients from cirrhotic patients, one in serum (miR-122, miR-885-5p, miR-221, miR-181b ) and one
[49]
in plasma (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-803 ). More recently, Tat
[61]
Trung et al. showed that a miRNA panel including miR-21, miR-122 and miR-192 had a good diagnostic
performance in discriminating HCC patients from other groups, in particular from cirrhotic and chronic
hepatitis patients.
Another potential limitation of several published studies is the fact that they were based on the assumption
that cell-free miRNA levels were altered as a consequence of their release by neoplastic cells, and tried to
validate as circulating cancer biomarkers the same miRNAs deregulated in tumor tissues. This assumption
may not be correct [62,63] . In fact, if we consider that ctDNA represents a small or very small fraction of
cfDNA (approximately 0.1%-1% or less for non-metastatic tumors and 1%-10% for large metastatic tumors),
it is difficult to conceive that cancer cells could instead release such a high amount of RNA to significantly
change content and levels of specific circulating RNAs. While this consideration raises doubts about
[64]
the source of circulating RNAs, a number of studies (see review by McAllister and Weinberg ) offer a
possible explanation by indicating that cancer should be considered a systemic disease. In this view, tumor-
associated systemic processes may unbalance the release of miRNAs from non-tumor cells and therefore
change circulating RNA profiles. Thus, finding altered circulating miRNA profiles would represent evidence
of a systemic pathophysiological process closely linked to the presence of a tumor, and such RNAs would
then represent actual circulating tumor biomarkers, although largely not directly released by tumor cells. In
this viewpoint, the numerous evidences that the altered levels of miRNAs in circulation do not necessarily
reflect the miRNA deregulation found in cancer tissues would become plausible and results should be re-