Page 104 - Read Online
P. 104
Sen. Hepatoma Res 2018;4:37 I http://dx.doi.org/10.20517/2394-5079.2018.39 Page 3 of 7
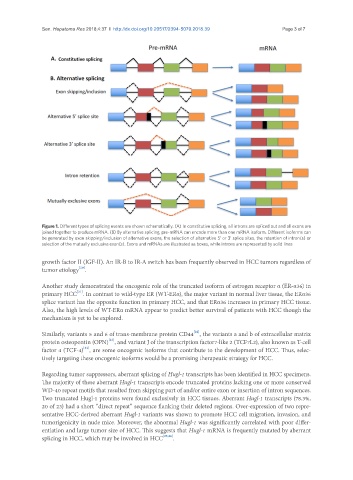
Figure 1. Different types of splicing events are shown schematically. (A) In constitutive splicing, all introns are spliced out and all exons are
joined together to produce mRNA. (B) By alternative splicing, pre-mRNA can encode more than one mRNA isoform. Different isoforms can
be generated by exon skipping/inclusion of alternative exons, the selection of alternative 5’ or 3’ splice sites, the retention of intron(s) or
selection of the mutually exclusive exon(s). Exons and mRNAs are illustrated as boxes, while introns are represented by solid lines
growth factor II (IGF-II). An IR-B to IR-A switch has been frequently observed in HCC tumors regardless of
[20]
tumor etiology .
Another study demonstrated the oncogenic role of the truncated isoform of estrogen receptor α (ER-α36) in
[21]
primary HCC . In contrast to wild-type ER (WT-ERα), the major variant in normal liver tissue, the ERα36
splice variant has the opposite function in primary HCC, and that ERα36 increases in primary HCC tissue.
Also, the high levels of WT-ERα mRNA appear to predict better survival of patients with HCC though the
mechanism is yet to be explored.
[22]
Similarly, variants 5 and 6 of trans-membrane protein CD44 , the variants a and b of extracellular matrix
[23]
protein osteopontin (OPN) , and variant J of the transcription factor7-like 2 (TCF7L2), also known as T-cell
[24]
factor 4 (TCF-4) , are some oncogenic isoforms that contribute to the development of HCC. Thus, selec-
tively targeting these oncogenic isoforms would be a promising therapeutic strategy for HCC.
Regarding tumor suppressors, aberrant splicing of Hugl-1 transcripts has been identified in HCC specimens.
The majority of these aberrant Hugl-1 transcripts encode truncated proteins lacking one or more conserved
WD-40 repeat motifs that resulted from skipping part of and/or entire exon or insertion of intron sequences.
Two truncated Hugl-1 proteins were found exclusively in HCC tissues. Aberrant Hugl-1 transcripts (78.3%,
20 of 23) had a short “direct repeat” sequence flanking their deleted regions. Over-expression of two repre-
sentative HCC-derived aberrant Hugl-1 variants was shown to promote HCC cell migration, invasion, and
tumorigenicity in nude mice. Moreover, the abnormal Hugl-1 was significantly correlated with poor differ-
entiation and large tumor size of HCC. This suggests that Hugl-1 mRNA is frequently mutated by aberrant
splicing in HCC, which may be involved in HCC [25,26] .